Industrial and Agricultural Waste Applied in the Development of Lithium-ion Batteries
Author(s)
Han-Sheng ChuangBiography
Prof. Han-Sheng Chuang is currently a full professor in the Department of Biomedical Engineering at National Cheng Kung University (NCKU), Taiwan. He was also jointly appointed Division Director of the Common Lab in the Center for Micro/Nano Science and Technology at NCKU.
Academy/University/Organization
National Cheng Kung UniversitySource
https://pubs.acs.org/doi/abs/10.1021/acssensors.8b00790
https://www.nature.com/articles/srep46336
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0148864-
TAGS
-
Share this article
You are free to share this article under the Attribution 4.0 International license
- ENGINEERING & TECHNOLOGIES
- Text & Image
- January 21,2020
The golden age brought by the advent of antibiotics since 1928 has faced an unprecedented challenge from the rapidly growing drug-resistant microbes (i.e., superbugs). The superbug spread has loomed as a global crisis and claims millions of lives every year. A gold standard, termed antimicrobial susceptibility testing (AST), has high effectiveness in the treatment of bacterial infections, but suffers from a lengthy turnaround time (3~5 days). Automated machines provide effective measures to save intensive human labor and enable parallel AST processing by optimizing time management. The turnaround time of each sample is reduced from days to around 1.5 days. Instead of tedious cell culture, unconventional approaches, such as multiplexed PCR and mass spectroscopy, claim to push the limit to an hour or even a few minutes. However, their minimum inhibitory concentrations remain undetermined. Thinking out of the box, a research team at National Cheng Kung University proposed a novel probe for ultrasensitive detection using Brownian motion. By appropriately modifying the surface of microbeads, each functionalized microbead can then act as a probe to interact with target bacteria and transmit the signal in terms of diffusion. Through investigating the trend patterns of the interactions between bacteria and microbeads with respect to bacterial species and drug types, the turnaround time of AST is greatly reduced to less than three hours. An accuracy of nearly 90% can be achieved as well. This novel technique appears to open up a new opportunity for better treatments of bacterial infections with precision medicine in the near future.
The first antibiotic, penicillin, was discovered and studied by Alexander Fleming in 1928. This achievement eventually brought him and another two scientists, Howard Flory and Ernst Boris Chain, a Nobel prize in medicine in 1945. After nine decades, however, the glory has worn out due to the rapidly growing drug-resistant microbes (i.e., superbugs) all around the world which have arisen because of antibiotics abuse. The emergence of superbugs is commonly attributed to three routes: hospitals, pharmacies, and animal husbandry (Fig. 1). Drug abuse is particularly serious on farms due to lack of awareness and knowledge. The spread of superbugs therefore looms as a global crisis and claims millions of lives every year.
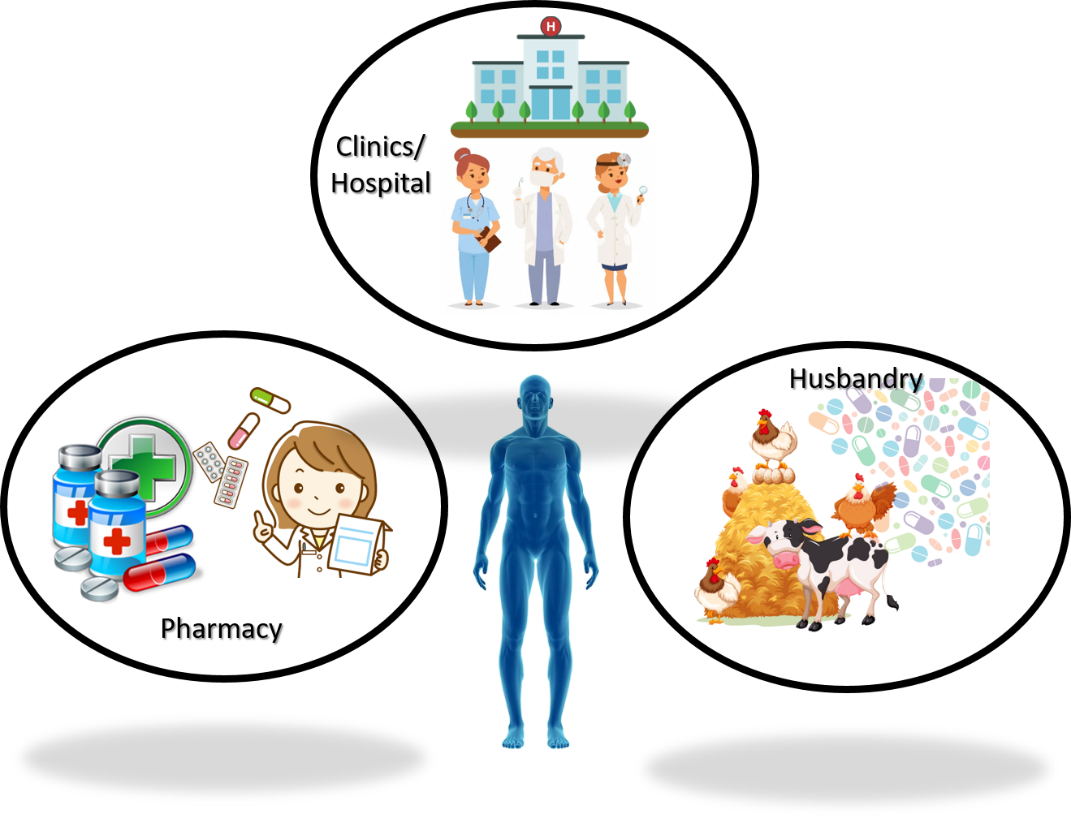
Figure 1. Three major routes that lead to the emergence of drug-resistant bacteria.
A gold standard of antibiotic administration to infections in clinics as of today remains dependent on a protocol, named antimicrobial susceptibility testing (AST). In spite of high effectiveness in the treatments of bacterial infections, the turnaround time of conventional AST requires 3~5 days (Fig. 2), which is insufficient to deal with some severe infections, such as sepsis. As a result, a rapid and precise antibiotic prescription is key to preventing the situation from deterioration. Notably, precision medicine has been emerging as a new hope for humans since it was initiated by U.S. president, Barack Obama, in 2015. By carefully seeking the right recipe for each individual, it is believed that the lid can be put back on Pandora’s box.
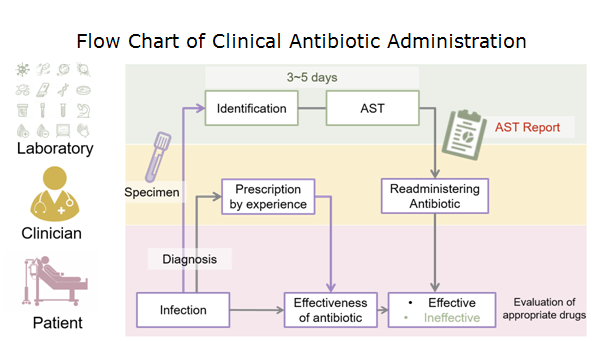
Figure 2. Flow chart of clinical antibiotic administration. The turnaround time of AST takes 3~5 days to generate a valid report.
The overall concept relies on developing a rapid AST to enable correct antibiotics to be administered to patients in a timely fashion, so that drug abuse can be mitigated in all aspects. To this end, numerous technologies have been developed to tackle the lengthy process of microbial culture in the current AST over the past decades. Automation is an effective measure to save intensive human labor and enable parallel AST processing by optimizing time management. The turnaround time of each sample is then reduced from 3~5 days to around 1.5 days. Major game players nowadays in the market, such as Vitek from BIOMÉRIEUX and Phoenix from BD, have successfully deployed their products to clinics and medical laboratories, hence revolutionizing the current treatments of bacterial infections. Lately, instead of culture, multiplexed PCR and an ultrafast DNA amplification named digital real-time loop-mediated isothermal amplification are claimed to push the limit to an hour or less in a minimal sample by identifying the drug-resistant bacteria from their genetic maps. Moreover, mass spectroscopy can even determine microbial species in a few minutes without tedious cell culture. Unfortunately, both methods are incapable of determining minimum inhibitory concentrations for antibiotics in real clinical treatments. Given that culture remains a necessary means to reveal the interactions between drugs and microbes, a novel technique utilizing sensitive probes to delineate the tiny kinetic change in the interaction was then proposed by a research group (https://bmoems.weebly.com/) in the Department of Biomedical Engineering at National Cheng Kung University, Taiwan. Through building up a database of temporal changes with respect to different bacteria species and drug types, the turnaround time of AST can be greatly reduced to less than three hours. A new probe suitable for such ultrasensitive detection mainly relies on the principle of Brownian motion. Brownian motion has long been misunderstood as useless random motion. However, a simple relation stated by Einstein is now imparting this natural phenomenon with a new meaning. With an appropriate surface modification on microbeads, each microbead can act as a probe to interact with target bacteria and transmit the signal in terms of diffusion (Fig. 3).
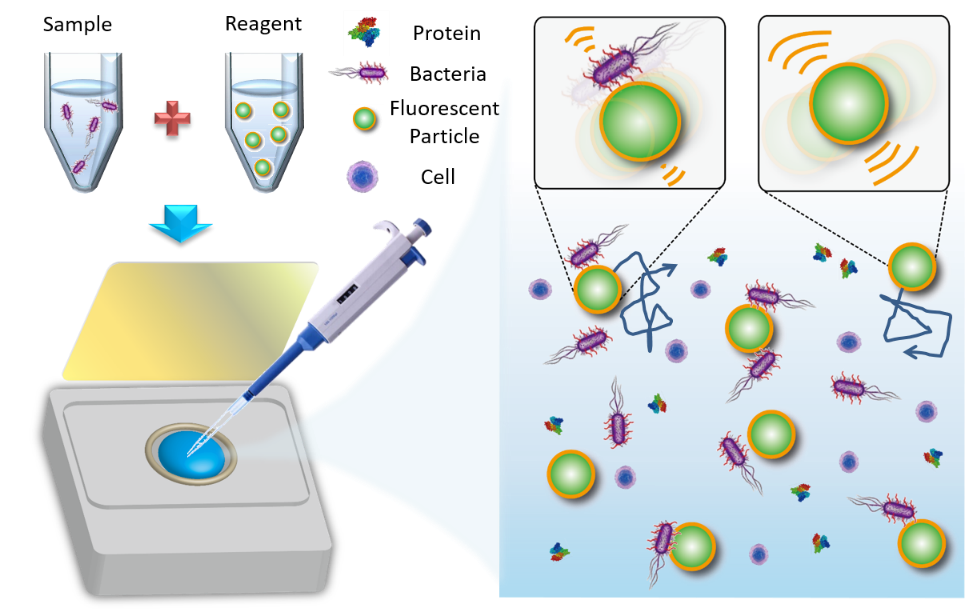
Figure 3 Schematic of the self-powered microbead sensors for monitoring of live microorganisms (left). Theoretical concept of diffusivity changes between functionalized microbeads with and without bacteria attached (right). In general, microbeads bound with bacteria express weaker Brownian motion.
The diffusivity, in general, is inversely proportional to the microbead size. When bacteria are resistant to an antibiotic, proliferative bacteria will bring down the diffusivity. Conversely, the diffusivity will tend to maintain a constant level over time as bacteria proliferation is inhibited by the drug. Now that only trend patterns are considered in this approach, a tedious cell culture can be avoided (Fig. 4). To counteract the background noise, reference microbeads (non-functional) are added in the same medium to eliminate the temperature, viscosity, and environmental vibration. A preclinical test with urine samples guided by the CLSI protocol has been conducted and showed good agreement with their conventional AST counterpart. An accuracy of nearly 90% was achieved. Considering that superbugs have been gradually posing a serious threat to human society, we are facing a turning point to take action before the situation becomes out of control. The aforementioned novel technique appears to open a new opportunity for better treatments of bacterial infections with precision medicine in the near future.
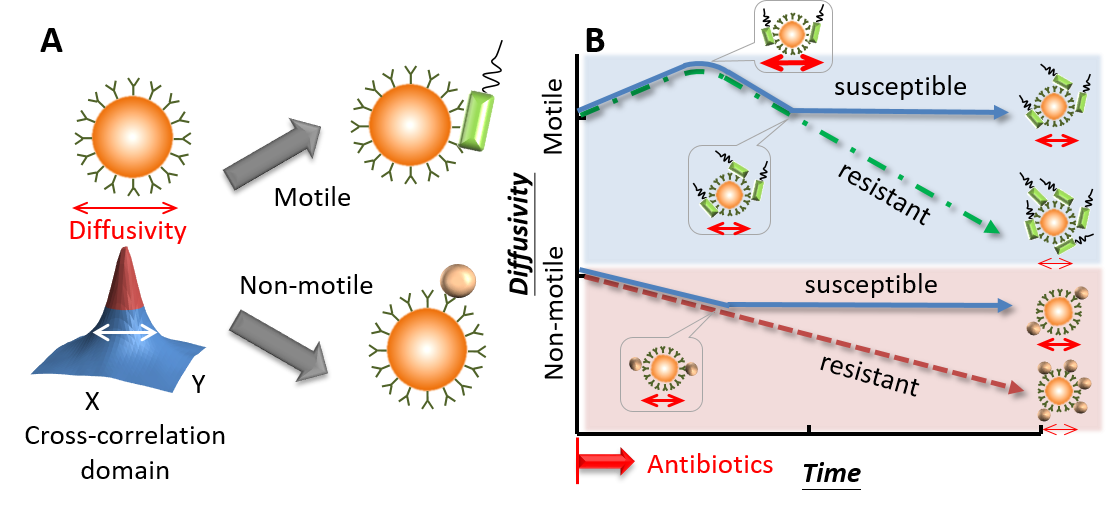
Figure 4. Conceptual illustration of the diffusometry-enabled technique for AST. (A) Fluorescent particles are measured by diffusometry and are analyzed with the spatial cross-correlation algorithm for the information of diffusivity. Theoretically, a broad correlation width indicates strong diffusivity. The target microorganisms are captured based on immunoreactions. Motile or non-motile bacteria can lead to different diffusivity changes. (B) In the AST tests, motile bacteria, such as P. aeruginosa, boost the diffusivity initially owing to the bacterial motility. After more bacteria attach to particles, the diffusivity drops all the way down if the bacteria are resistant to antibiotics; otherwise, the diffusivity stops declining at a certain time point. By contrast, non-motile bacteria have no increased diffusivity over time. Apart from this difference, their other responses to antibiotics are similar to their motile counterparts.
STAY CONNECTED. SUBSCRIBE TO OUR NEWSLETTER.
Add your information below to receive daily updates.